Igv Number of Reads Aligned to Location
Alignment to Read Counts & Visualization in IGV
This document assumes preproc htstream has been completed.
IF for some reason it didn't finish, is corrupted or you missed the session, y'all can re-create over a completed copy
cp -r /share/biocore/workshops/2019_March_RNAseq/HTS_testing /share/workshop/$USER/rnaseq_example/. cp -r /share/biocore/workshops/2019_March_RNAseq/01-HTS_Preproc /share/workshop/$USER/rnaseq_example/. cp /share/biocore/workshops/2019_March_RNAseq/summary_hts.txt /share/workshop/$USER/rnaseq_example/. Alignment vs Assembly
Given sequence information,
Assembly seeks to put together the puzzle without knowing what the motion picture is.
Mapping tries to put together the puzzle pieces straight onto an paradigm of the picture.
In mapping the question is more, given a pocket-size chunk of sequence, where in the genome did this piece most likely come from.
The goal and so is to find the lucifer(es) with either the "best" edit altitude (smallest), or all matches with edit distance less than max edit dist. Main issues are:
- Large search infinite
- Regions of similarity (aka repeats)
- Gaps (INDELS)
- Complexity (RNA, transcripts)
Considerations
- Placing reads in regions that do not be in the reference genome (reads extend off the finish) [ mitochondrial, plasmids, structural variants, etc.].
- Sequencing errors and variations: alignment betwixt read and truthful source in genome may have more differences than alignment with some other re-create of repeat.
- What if the closest fully sequenced genome is also divergent? (3% is a common alignment capability)
- Placing reads in repetitive regions: Some algorithms only return 1 mapping; If multiple: map quality = 0
- Algorithms that use paired-end information => might prefer correct distance over correct alignment.
In RNAseq data, you must also consider effect of splice junctions, reads may span an intron.
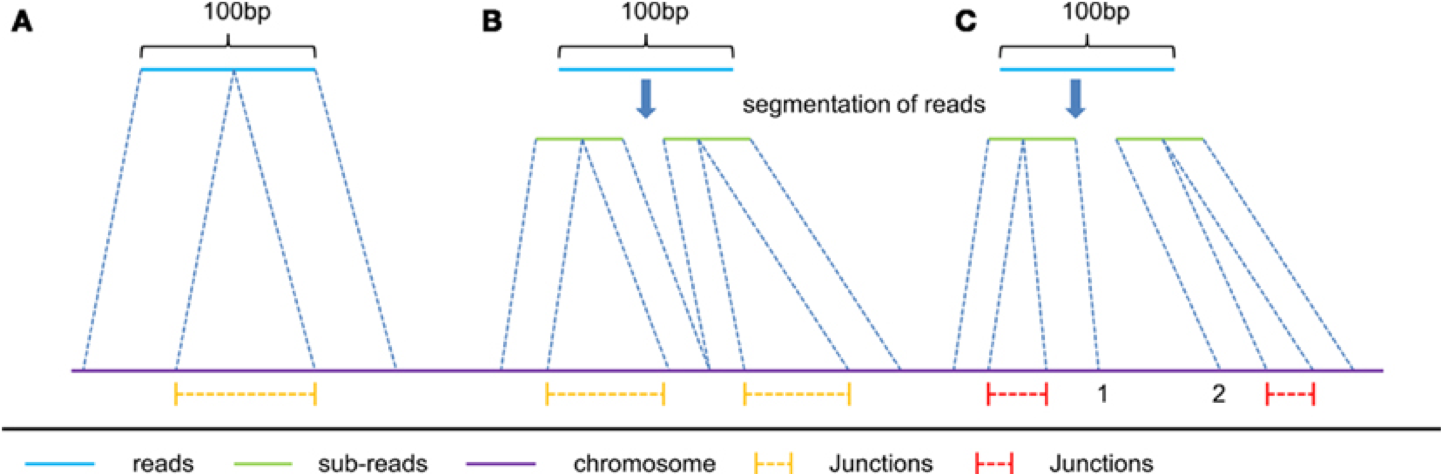
Aligners
Many alignment algorithms to cull from.
- Spliced Aligners
- STAR
- HiSAT2 (formerly Tophat [Bowtie2])
- GMAP - GSNAP
- SOAPsplice
- MapSplice
- Aligners that tin 'clip'
- bwa-mem
- Bowtie2 in local way
Pseudo-aligners (salmon and kalisto)
- Quasi-mapping
- Probabilistic
- Map to transcripts, not genome
- Does transcript quantifications (or gene)
- Blazing FAST and tin can run on nigh laptops
- Experience suggests differences between "traditional" mappers are in the low affluence genes.
Mapping against the genome vs transcriptome
- May seem intuitive to map RNAseq data to transcriptome, but it is not that unproblematic.
- Transcriptomes are rarely complete.
- Which transcript of a gene should you map to, canonical transcript?
- Shouldn't map to all splice variants as these would show up as 'multi-mappers'.
- More so, a mapper will try to map every read, somewhere, provided the result meets its minimum requirements.
- Need to provide a mapper with all possible places the read could accept arisen from, which is best represented past the genome.
- Otherwise, you lot get mis-mapping because its close enough.
Genome and Genome Note
Genome sequence fasta files and note (gff, gtf) files go together! These should be identified at the beginning of analysis.
- Genome fasta files should include all primary chromosomes, unplaced sequences and un-localized sequences, also as any organelles. Should bet contain any contigs that represent patches, or culling haplotypes.
- If y'all expect contamination, or the presence of additional sequence/genome, add the sequence(s) to the genome fasta file.
- Annotation file should be GTF (preferred), and should exist the most comprehensive you tin observe.
- Chromosome names in the GTF must match those in the fasta file (they don't always do).
- Star recommends the Genecode annotations for mouse/man
Counting reads as a proxy for factor expression
The more you can count (and HTS sequencing systems can count a lot) the better the measure of copy number for even rare transcripts in a population.
- Most RNA-seq techniques deal with count information. Reads are mapped to a reference genome, transcripts are detected, and the number of reads that map to a transcript (or factor) are counted.
- Read counts for a transcript are roughly proportional to the factor'south length and transcript affluence (in whole transcript methods).
Technical artifacts should be considered during counting
- Mapping quality
- Map-ability (uniqueness), the read is not ambiguous
Options are (STAR, HTSEQ, featureCounts)
The HTSEQ mode
Trouble:
- Given a sam/bam file with aligned sequence reads and a list of genomic feature (genes locations), nosotros wish to count the number of reads (fragments) than overlap each characteristic.
- Features are divers by intervals, they have a showtime and stop position on a chromosome.
- For this workshop and analysis, features are genes which are the union of all its exons. You could consider each exon as a characteristic, for culling splicing.
- Htseq-count has iii overlapping modes
- spousal relationship
- intersection-strict
- intersection-nonempty

- from the htseq paper
Star Implementation
Counts coincide with Htseq-counts under default parameters (matrimony and tests all orientations). Need to specify GTF file at genome generation step or during mapping.
- Output, 4 columns
- GeneID
- Counts for unstranded
- Counts for first read strand
- Counts for second read strand
Choose the appropriate cavalcade given the library preparation characteristics and generate a matrix table, columns are samples, rows are genes.
Alignment concepts
Indexing a Reference sequence and notation
1. Starting time lets brand sure we are where nosotros are supposed to be and that the References directory is available.
cd /share/workshop/$USER/rnaseq_example 2. To align our data we will demand the genome (fasta) and annotation (gtf) for man. There are many places to discover them, only we are going to become them from the GENCODE.
We need to offset become the url for the annotation gtf. For RNAseq nosotros want to use the PRI (primary) genome chromosome and corresponding annotation.
We need to first get the url for the genome fasta.
2. We are going to use an aligner called 'STAR' to align the information, but first we demand to index the genome for STAR. Lets pull down a slurm script to index the homo GENCODE version of the genome.
wget https://raw.githubusercontent.com/ucdavis-bioinformatics-grooming/2019_March_UCSF_mRNAseq_Workshop/principal/scripts/star_index.slurm less star_index.slurm When yous are washed, type "q" to exit.
#!/bin/bash #SBATCH --job-proper name=star_index # Job name #SBATCH --nodes=1 #SBATCH --ntasks=eight #SBATCH --time=120 #SBATCH --mem=40000 # Memory puddle for all cores (come across likewise --mem-per-cpu) #SBATCH --partition=product #SBATCH --reservation=workshop #SBATCH --account=workshop #SBATCH --output=slurmout/star-index_%A.out # File to which STDOUT volition be written #SBATCH --error=slurmout/star-index_%A.err # File to which STDERR will be written start = `date +%due south` echo $HOSTNAME outpath = "References" [[ -d ${ outpath } ]] || mkdir ${ outpath } cd ${ outpath } wget ftp://ftp.ebi.ac.great britain/pub/databases/gencode/Gencode_human/release_29/GRCh38.primary_assembly.genome.fa.gz gunzip GRCh38.primary_assembly.genome.fa.gz FASTA = "../GRCh38.primary_assembly.genome.fa" wget ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_29/gencode.v29.primary_assembly.annotation.gtf.gz gunzip gencode.v29.primary_assembly.annotation.gtf.gz GTF = "../gencode.v29.primary_assembly.annotation.gtf" mkdir star.overlap100.gencode.v29 cd star.overlap100.gencode.v29 module load star/2.7.0e call = "STAR --runThreadN 8 \ --runMode genomeGenerate \ --genomeDir . \ --sjdbOverhang 100 \ --sjdbGTFfile ${ GTF } \ --genomeFastaFiles ${ FASTA } " echo $telephone call eval $phone call end = `date +%south` runtime = $((cease-start)) echo $runtime - The script uses wget to download the fasta and GTF files from GENCODE using the links you constitute earlier.
- Uncompresses them using gunzip.
- Creates the star alphabetize directory [star.overlap100.gencode.v29].
- Alter directory into the new star index directory. we run the star indexing command from within the directory, for some reason star fails if you lot attempt to run it exterior this directory.
-
Run star in mode genomeGenerate.
sbatch star_index.slurm
This step will take a couple hours. You tin look at the STAR documentation while you lot wait. All of the output files will be written to the star_index directory.
IF for some reason it didn't finish, is corrupted, or yous missed the session, you tin copy over a completed copy.
cp -r /share/biocore/workshops/2019_March_RNAseq/References/star.overlap100.gencode.v29 /share/workshop/$USER/rnaseq_example/References/. Alignments
one. Nosotros are now set to try an alignment. Allow's create an output directory for STAR:
cd /share/workshop/$USER/rnaseq_example/HTS_testing and allow's run STAR (via srun) on the pair of streamed test files we created earlier. The command is on multiple lines for readability:
srun --time=xv:00:00 -n 8 --mem=32g --reservation=workshop --account=workshop --pty /bin/bash In one case you've been given an interactive session we can run STAR. Yous can ignore the two warnings/errors and you know your on a cluster node considering your server will alter. Hither you encounter I'm on tadpole, then after the srun command is successful, I am now on drove-13.
# msettles@polliwog:/share/workshop/msettles/rnaseq_example/> HTS_testing$ srun --time=15:00:00 -n 8 --mem=32g --> > reservation=workshop --account=workshop --pty /bin/bash # srun: chore 29372920 queued and waiting for resources # srun: task 29372920 has been allocated resource # groups: cannot discover name for grouping ID 2020 # bash: /domicile/msettles/.bashrc: Permission denied # msettles@drove-13:/share/workshop/msettles/rnaseq_example/> HTS_testing$ So run the star commands
module load star/2.7.0e STAR \ --runThreadN 8 \ --genomeDir ../References/star.overlap100.gencode.v29 \ --outSAMtype BAM SortedByCoordinate \ --quantMode GeneCounts \ --outFileNamePrefix SampleAC1.streamed_ \ --readFilesCommand zcat \ --readFilesIn SampleAC1.streamed_R1.fastq.gz SampleAC1.streamed_R2.fastq.gz In the command, nosotros are telling star to count reads on a gene level ('–quantMode GeneCounts'), the prefix for all the output files will be SampleAC1.streamed_, the control to unzip the files (zcat), and finally, the input file pair.
Once finished please 'get out' the srun session. You lot'll know you lot were successful when your back on tadpole
# msettles@drove-13:/share/workshop/msettles/rnaseq_example/HTS_testing$ go out # get out # msettles@tadpole:/share/workshop/msettles/rnaseq_example/HTS_testing$ At present permit'south take a look at an alignment in IGV.
1. We first need to index the bam file, will apply 'samtools' for this step, which is a programme to dispense SAM/BAM files. Accept a look at the options for samtools and 'samtools index'.
module load samtools/one.9 samtools samtools index Nosotros need to alphabetize the BAM file:
cd /share/workshop/$USER/rnaseq_example/HTS_testing samtools index SampleAC1.streamed_Aligned.sortedByCoord.out.bam IF for some reason information technology didn't finish, is corrupted or you lot missed the session, you can copy over a completed copy
cp -r /share/biocore/workshops/2019_March_RNAseq/HTS_testing/SampleAC1.streamed_Aligned.sortedByCoord.out.bam* /share/workshop/$USER/rnaseq_example/HTS_testing 2. Transfer SampleAC1.streamed_Aligned.sortedByCoord.out.bam and SampleAC1.streamed_Aligned.sortedByCoord.out.bam (the alphabetize file) to your computer using scp or winSCP, or copy/paste from cat [sometimes doesn't work].
In a new shell session on my laptop. NOT logged into tadpole.
mkdir ~/rnaseq_workshop cd ~/rnaseq_workshop scp msettles@tadpole.genomecenter.ucdavis.edu:/share/workshop/msettles/rnaseq_example/HTS_testing/SampleAC1.streamed_Aligned.sortedByCoord.out.bam* . Its ok of the mkdir control fails ("File exists") because we aleady created the directory earlier.
3. Now nosotros are ready to employ IGV. Go to the IGV folio at the Broad Constitute.
And then navigate to the download folio, IGV download 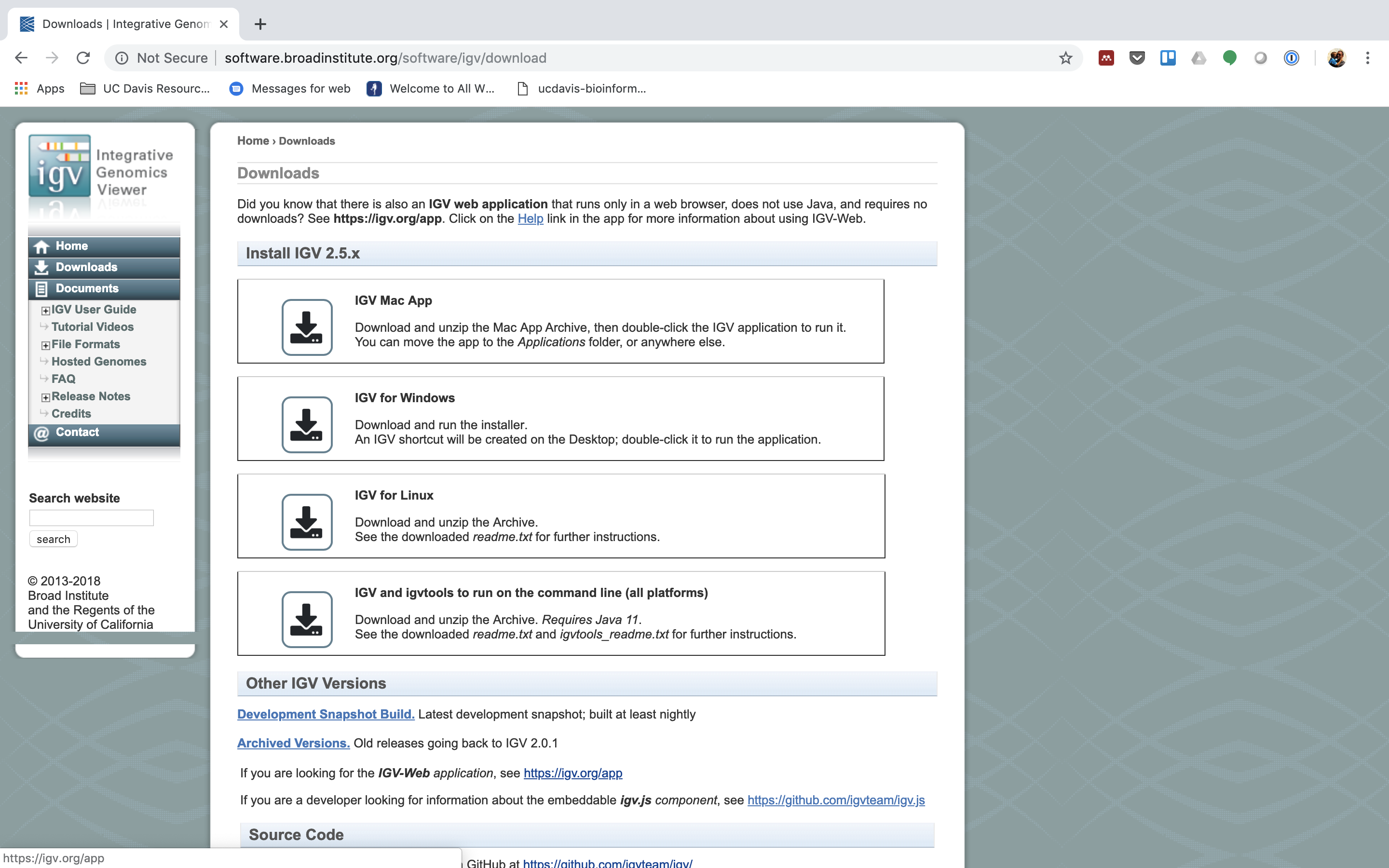
Here you tin can download IGV for your corresponding platform (Window, Mac OSX, Linux), but we are going to use the web application they supply, IGV web app.
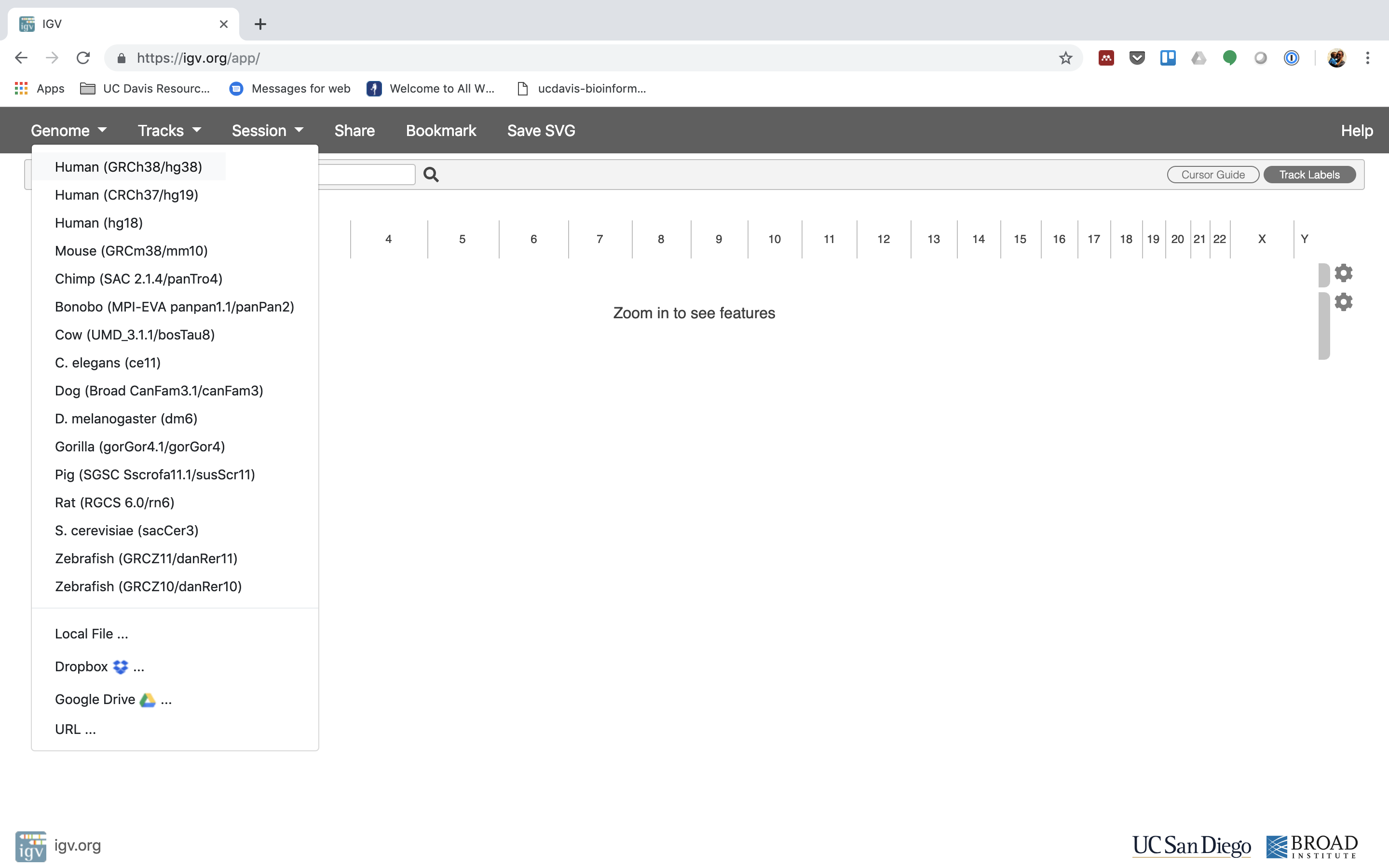
iv. The first thing nosotros want to do is load the Homo genome. Click on "Genomes" in the card and cull "Human (GRCh38/hg38)".
5. Now let's load the alignment bam and index files. Click on "Tracks" and cull "Local File …".
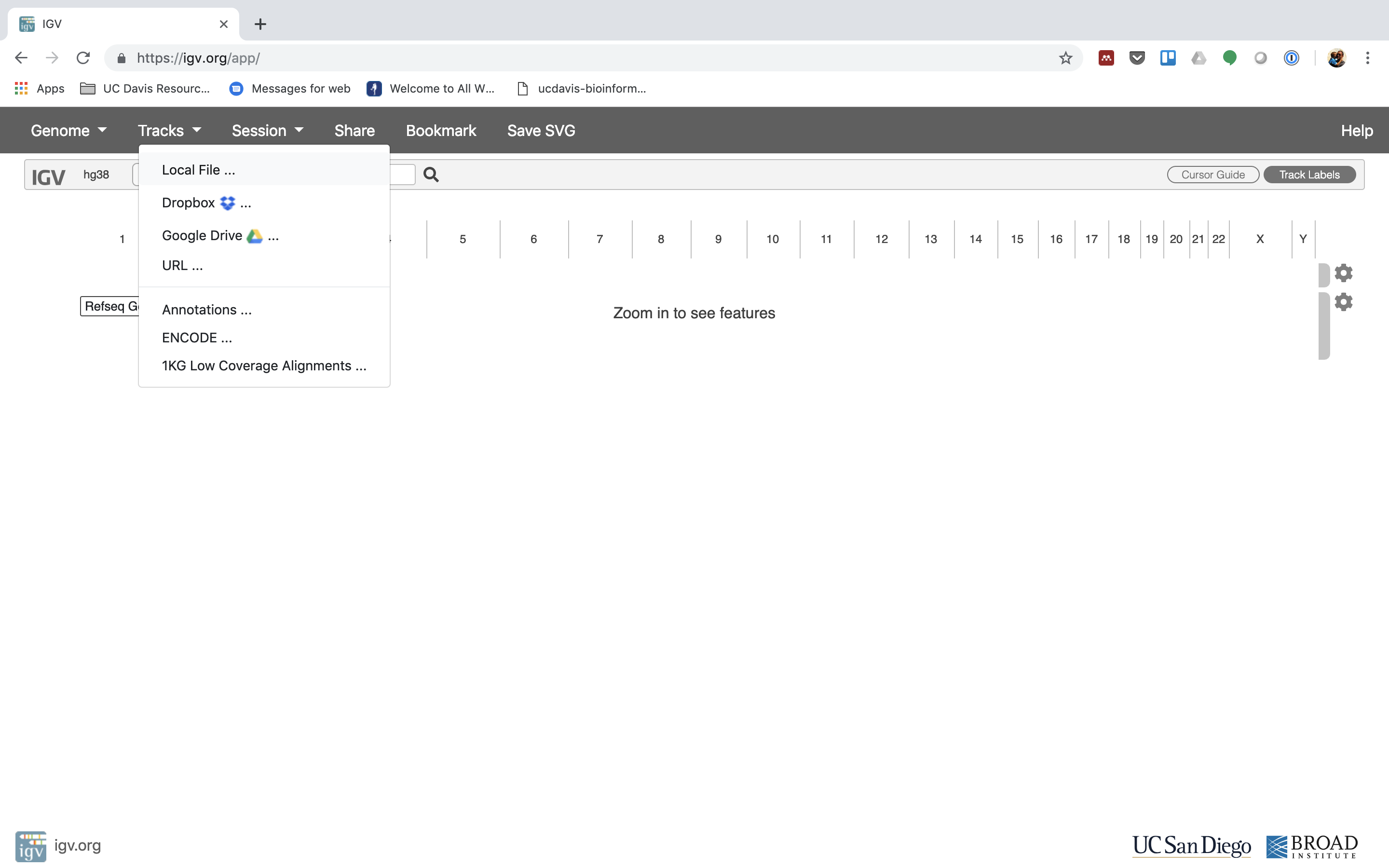
Navigate to where you transferred the bam and alphabetize file and select them both.
At present your alignment is loaded. Any loaded bam file aligned to a genome is called a "track".
6. Lets take a look at the alignment associated with the gene HBB:
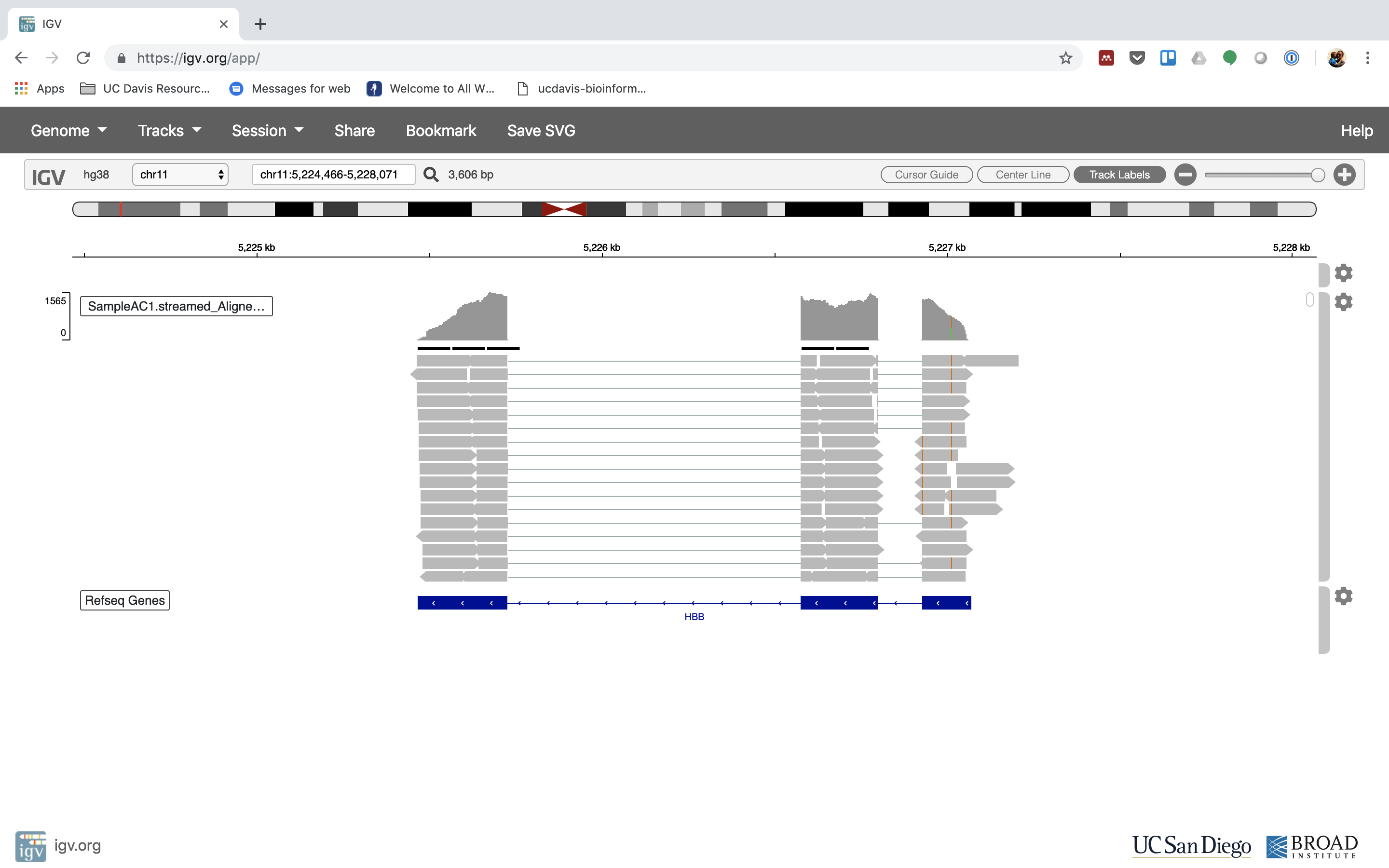
Yous can zoom in by clicking on the plus sign (summit right) or zoom out by clicking the negative sign. Y'all also may accept to motion around by clicking and dragging in the BAM track window.
Yous can also zoom in past clicking and dragging across the number line at the top. That section will highlight, and when you lot release the button, it will zoom into that department.
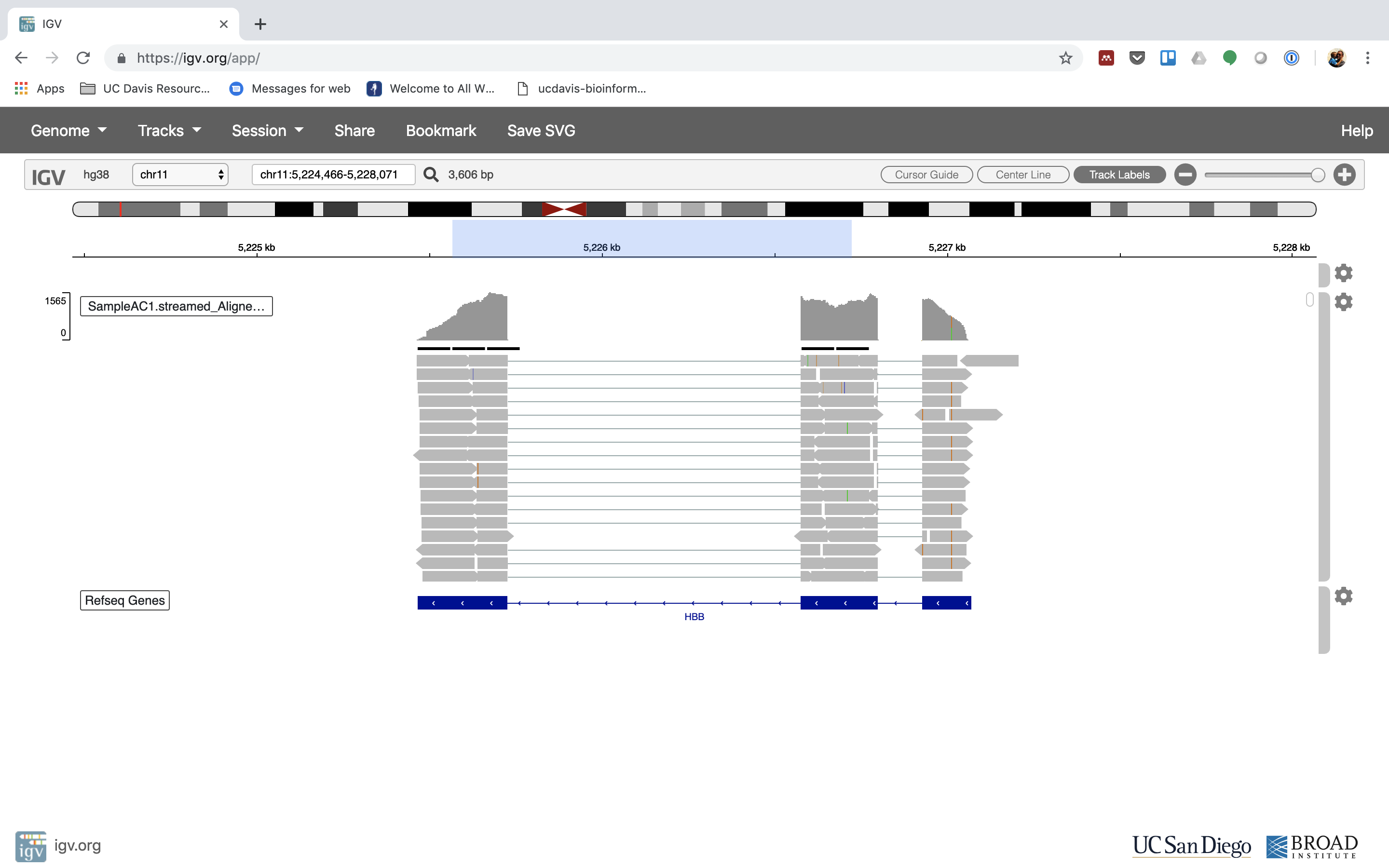
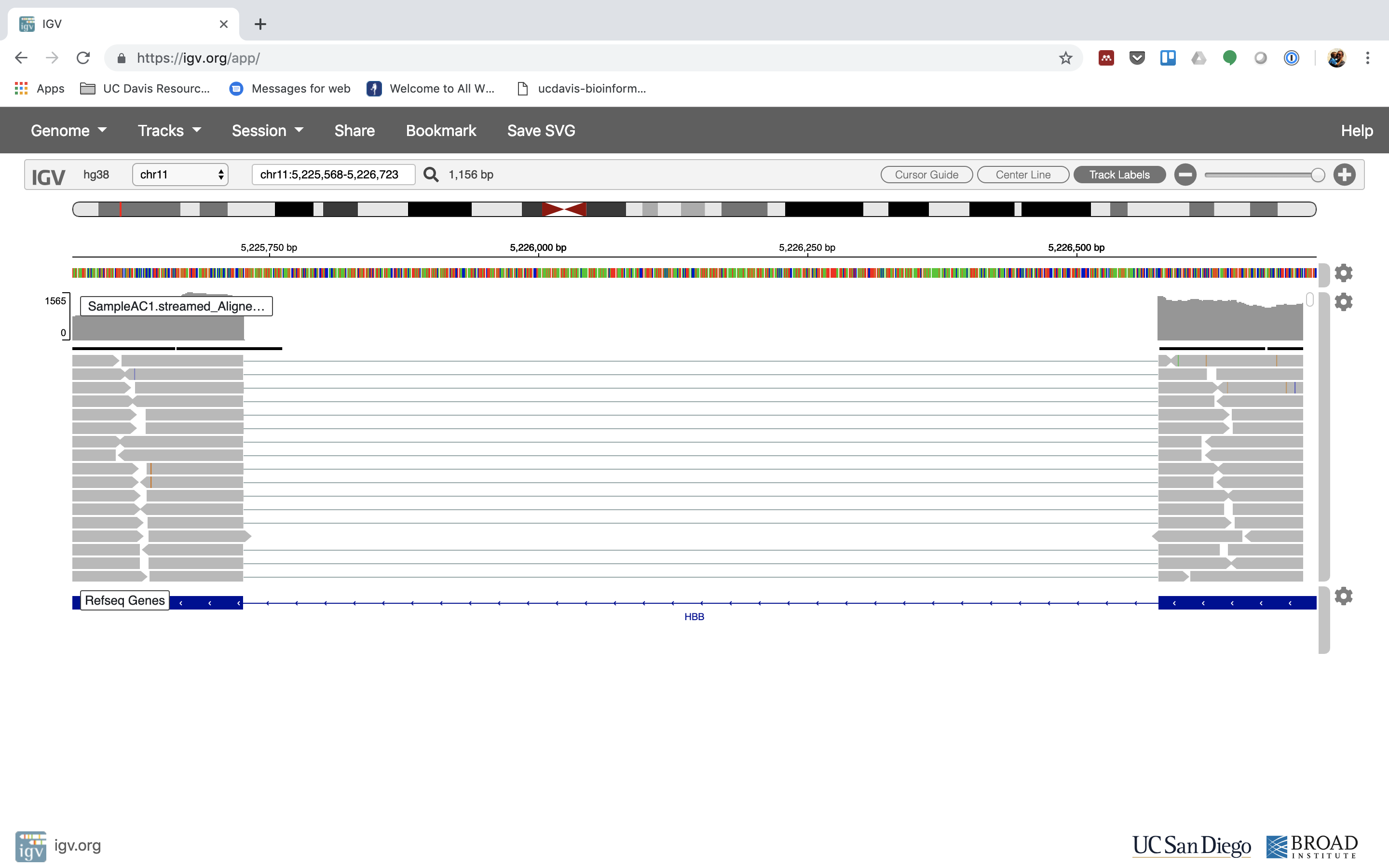
Reset the window by searching for HBB over again. And zoom in 1 step.
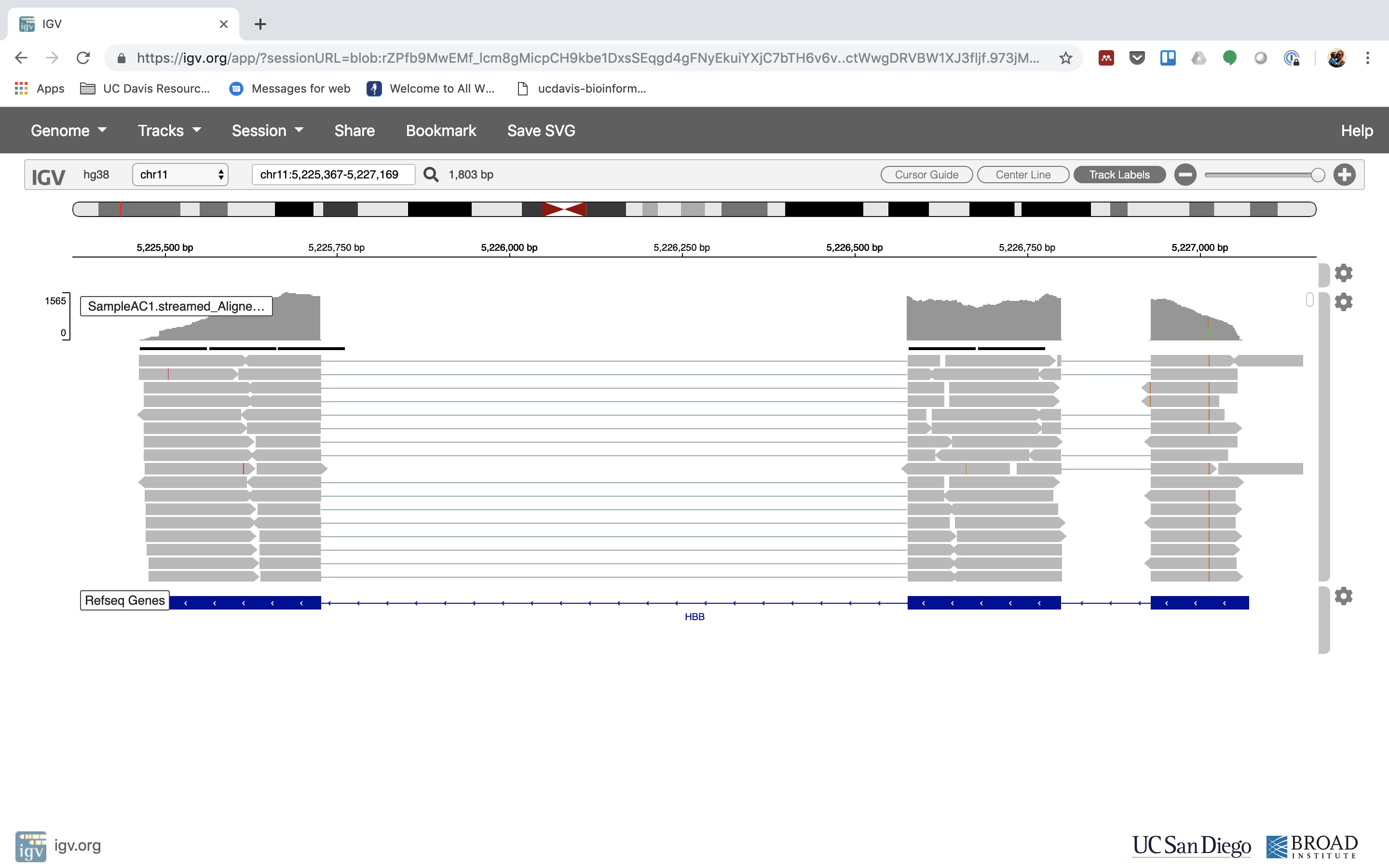
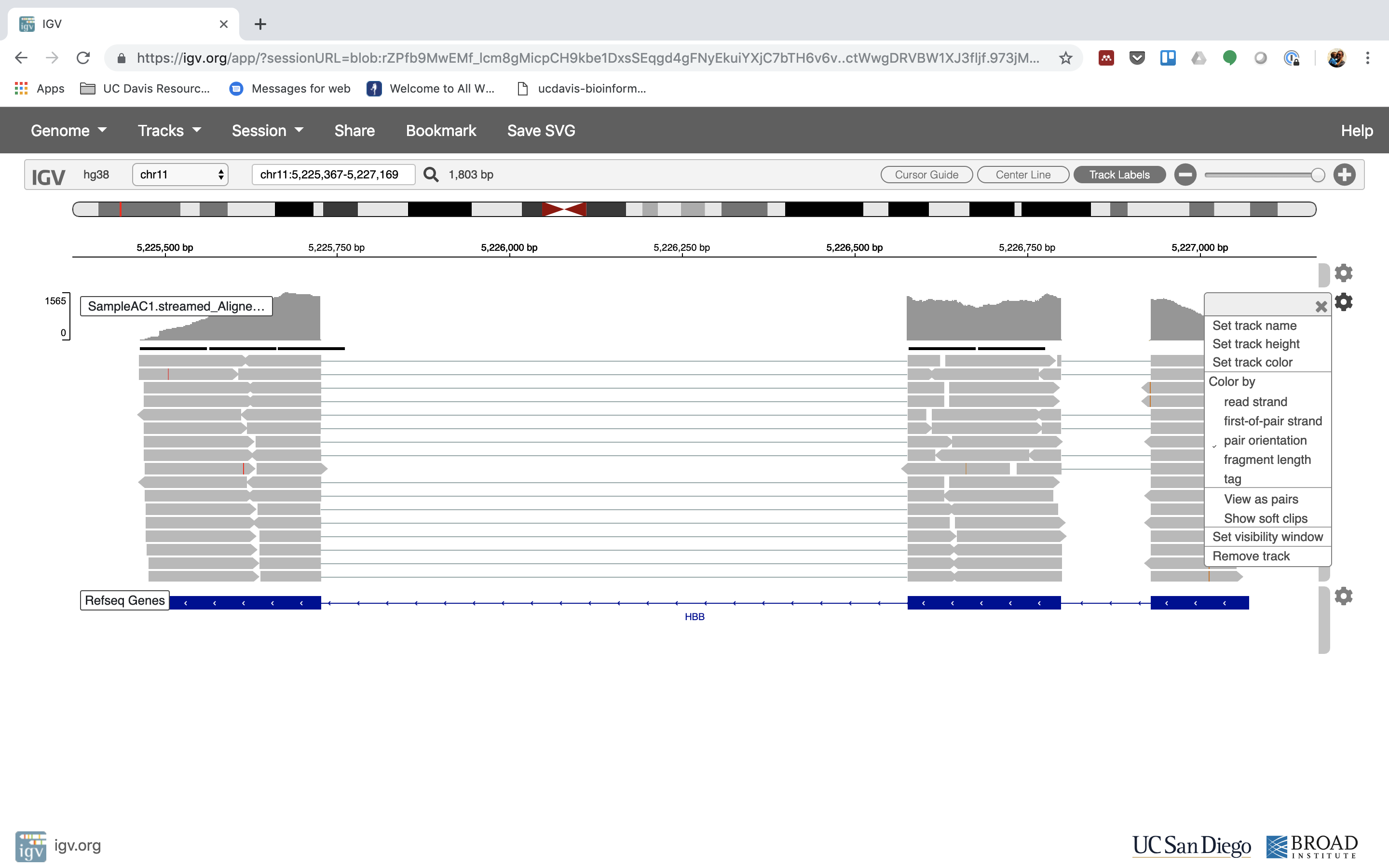
7. See that the reads should be adjustment within the exons in the gene. This makes sense, since RNA-Seq reads are from exons. Play with the settings on the right paw side a bit.
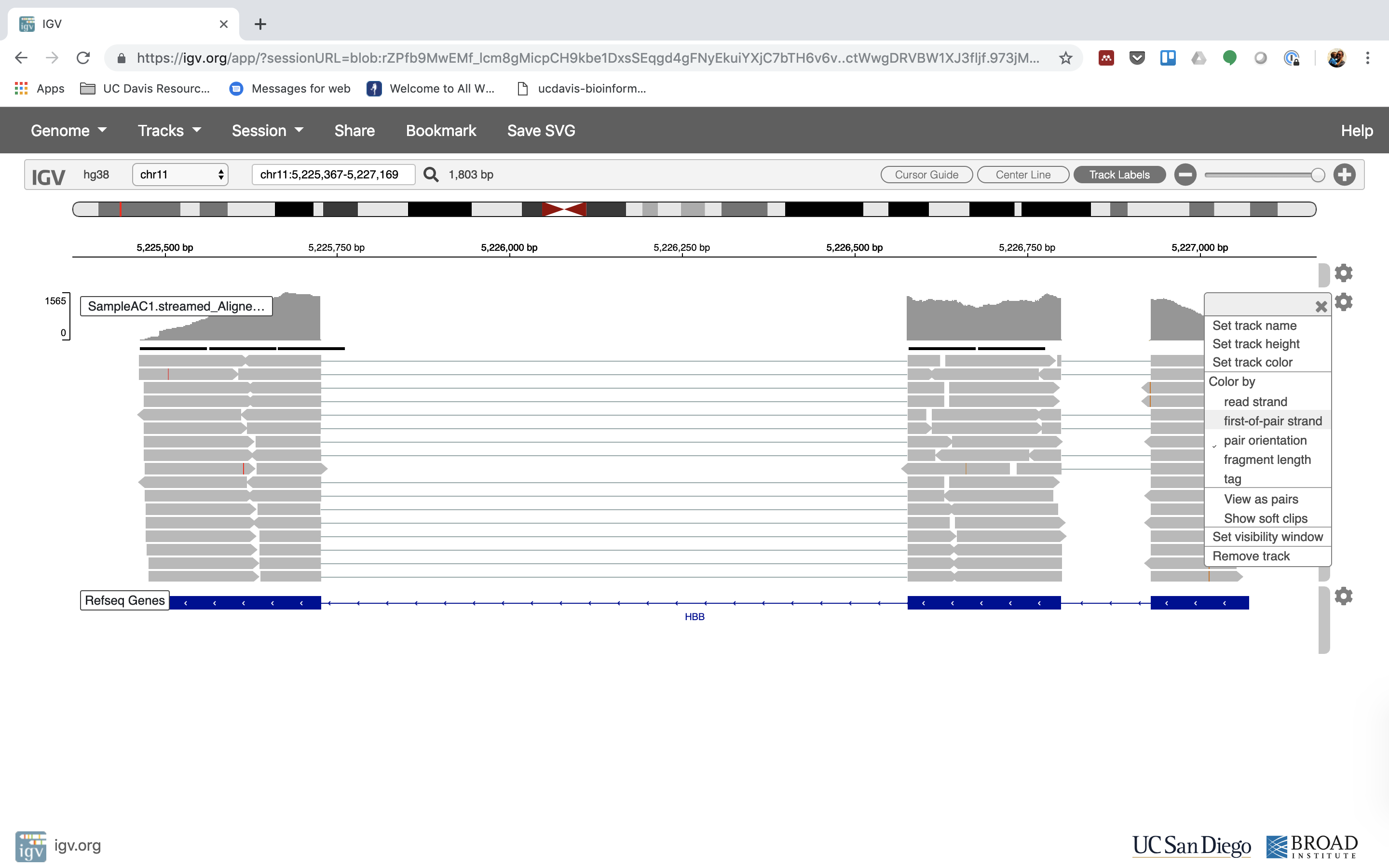
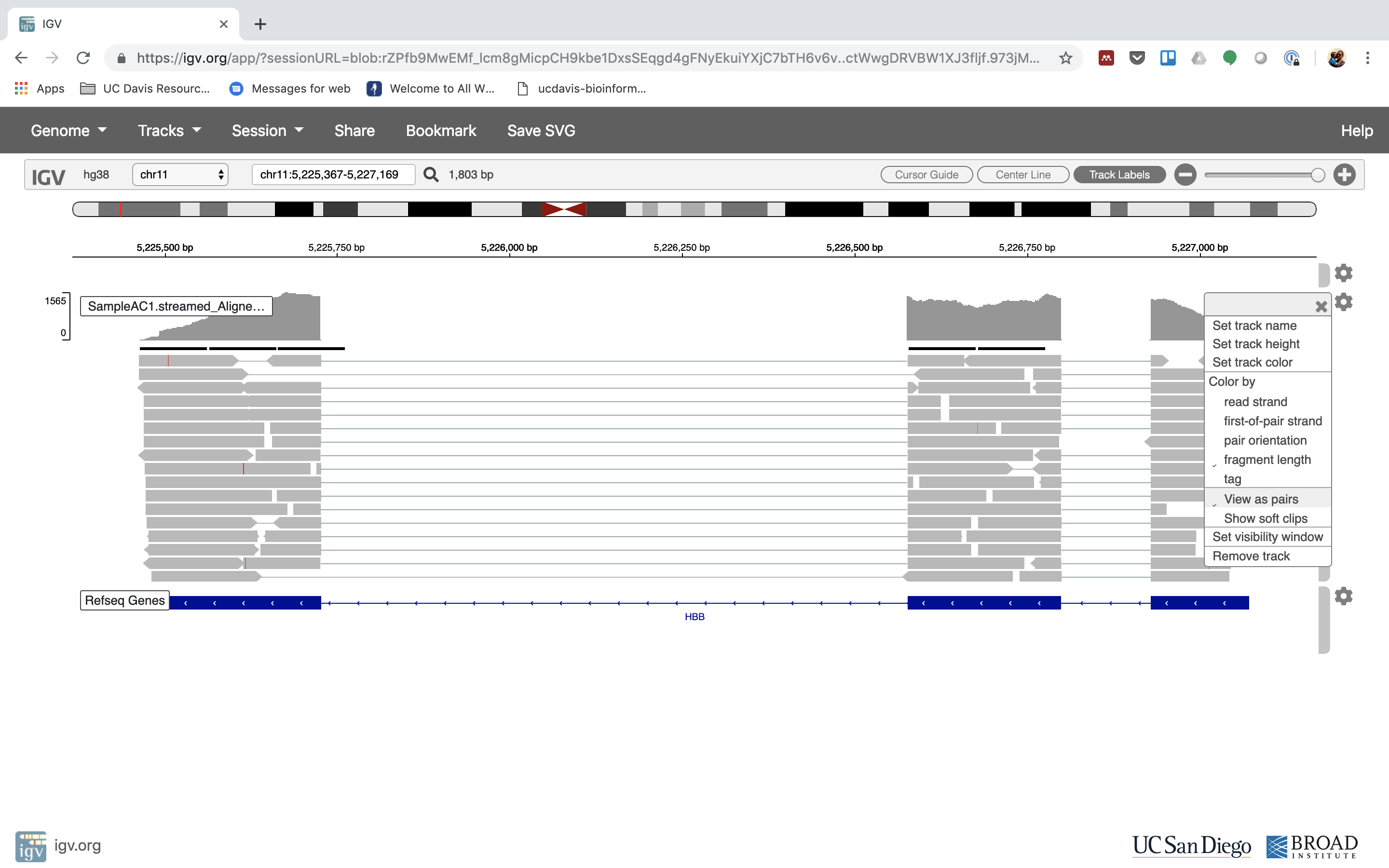
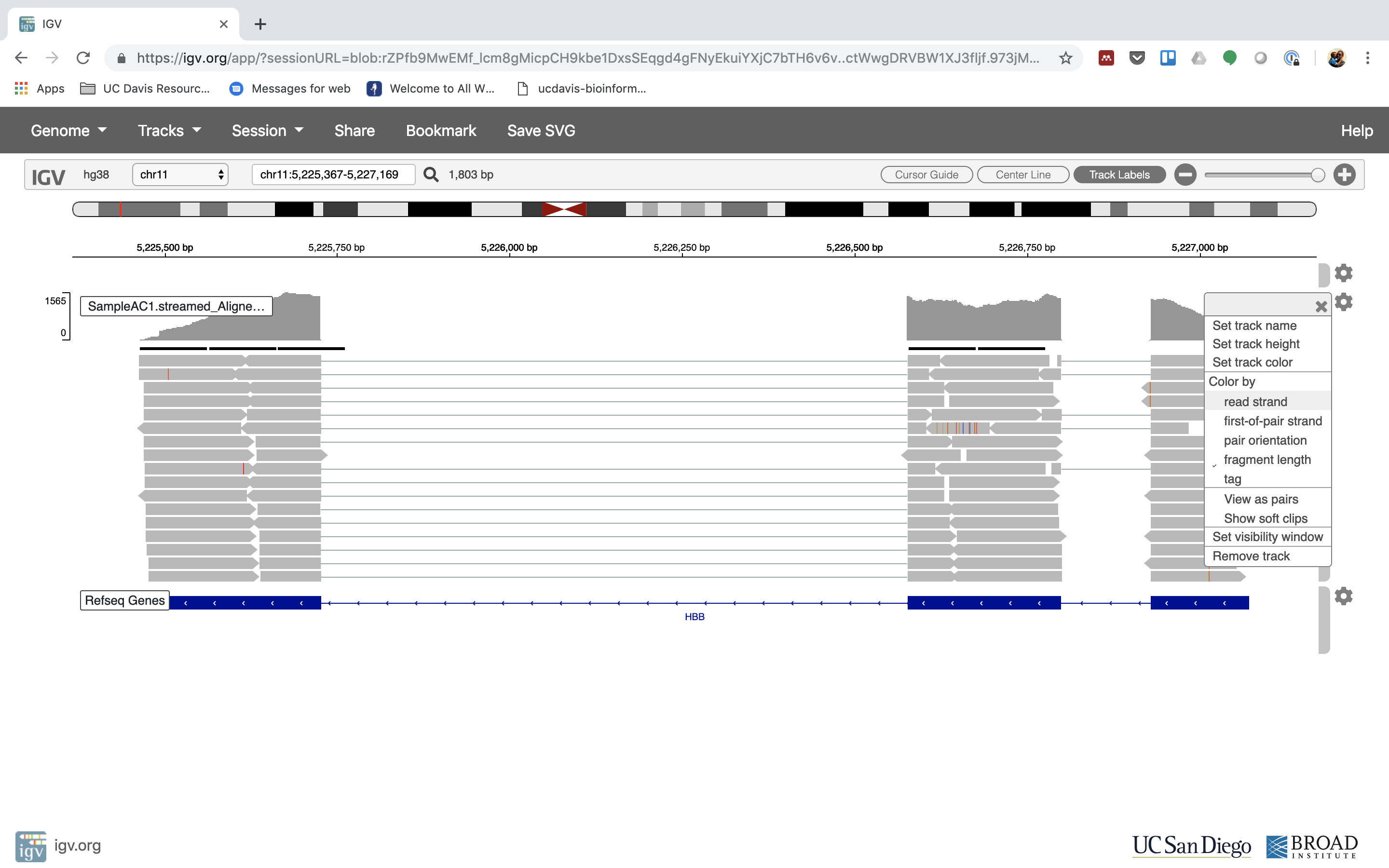
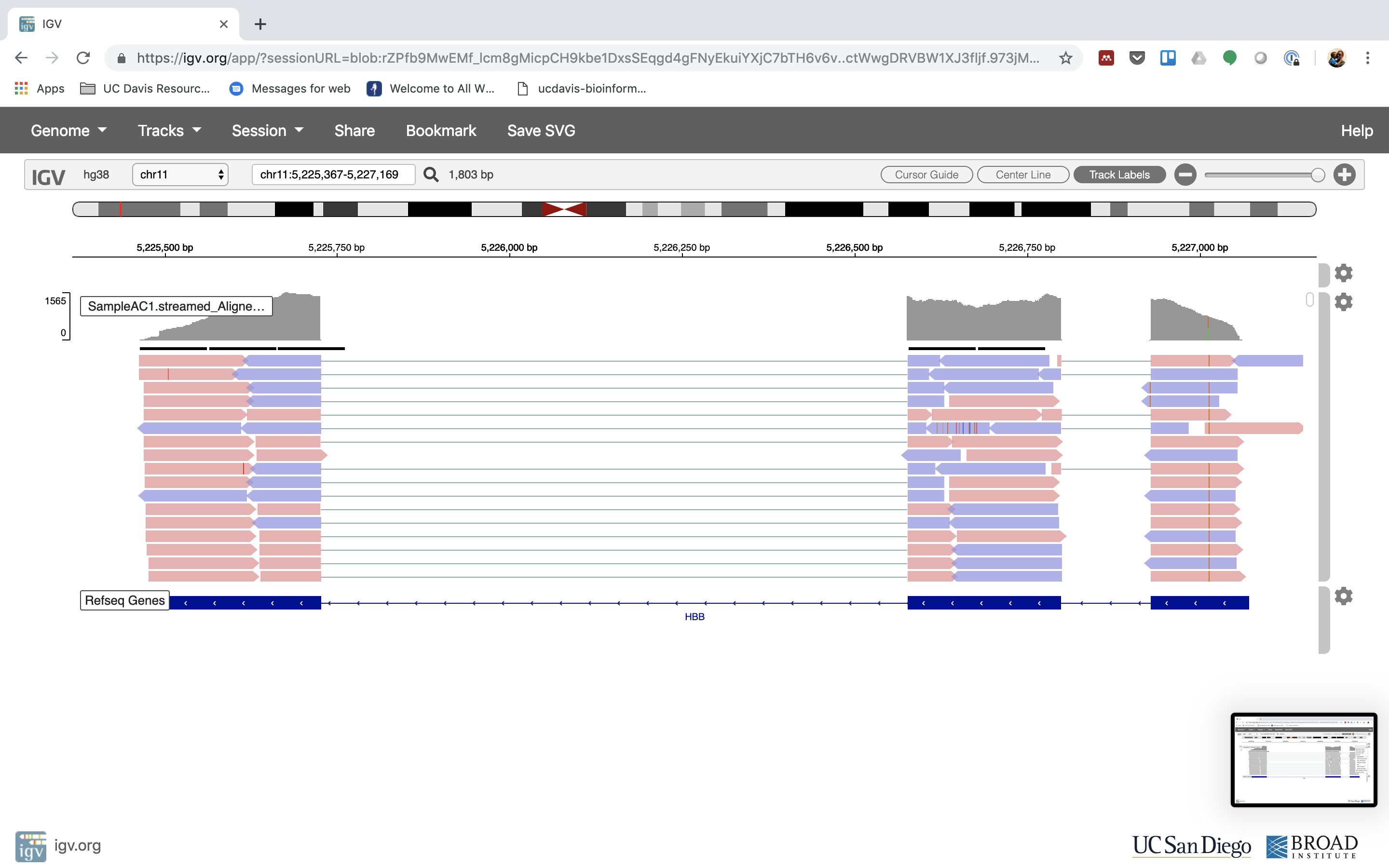
Running STAR on the experiment
1. We tin now run STAR across all samples on the real data using a SLURM script, star.slurm, that we should accept a look at now.
cd /share/workshop/$USER/rnaseq_example # Nosotros'll run this from the primary directory wget https://raw.githubusercontent.com/ucdavis-bioinformatics-training/2019_March_UCSF_mRNAseq_Workshop/main/scripts/star.slurm less star.slurm When you are done, type "q" to exit.
#!/bin/bash #SBATCH --task-proper noun=star # Job proper name #SBATCH --nodes=i #SBATCH --ntasks=8 #SBATCH --fourth dimension=threescore #SBATCH --mem=32000 # Memory puddle for all cores (run across also --mem-per-cpu) #SBATCH --sectionalisation=product #SBATCH --reservation=workshop #SBATCH --account=workshop #SBATCH --assortment=1-xvi #SBATCH --output=slurmout/star_%A_%a.out # File to which STDOUT will be written #SBATCH --error=slurmout/star_%A_%a.err # File to which STDERR will be written start = `date +%s` echo $HOSTNAME echo "My SLURM_ARRAY_TASK_ID: " $SLURM_ARRAY_TASK_ID sample = `sed " ${ SLURM_ARRAY_TASK_ID } q;d" samples.txt` REF = "References/star.overlap100.gencode.v29" outpath = '02-STAR_alignment' [[ -d ${ outpath } ]] || mkdir ${ outpath } [[ -d ${ outpath }/${ sample } ]] || mkdir ${ outpath }/${ sample } echo "SAMPLE: ${ sample } " module load star/ii.7.0e call = "STAR --runThreadN 8 \ --genomeDir $REF \ --outSAMtype BAM SortedByCoordinate \ --readFilesCommand zcat \ --readFilesIn 01-HTS_Preproc/ ${ sample } / ${ sample } _R1.fastq.gz 01-HTS_Preproc/ ${ sample } / ${ sample } _R2.fastq.gz \ --quantMode GeneCounts \ --outFileNamePrefix ${ outpath } / ${ sample } / ${ sample } _ \ ${ outpath } / ${ sample } / ${ sample } -STAR.stdout 2> ${ outpath } / ${ sample } / ${ sample } -STAR.stderr" echo $telephone call eval $call end = `date +%south` runtime = $((end-start)) echo $runtime Later looking at the script, lets run information technology.
sbatch star.slurm # moment of truth! We can watch the progress of our task array using the 'squeue' control. Takes about thirty minutes to process each sample.
squeue -u msettles # use your username Quality Balls - Mapping statistics as QA/QC.
one. Once your jobs have finished successfully (check the error and out logs similar we did in the previous exercise), use a script of ours, star_stats.sh to collect the alignment stats. Don't worry about the script's contents at the moment; you lot'll use very like commands to create a counts tabular array in the next section. For now:
cd /share/workshop/$USER/rnaseq_example # We'll run this from the chief directory wget https://raw.githubusercontent.com/ucdavis-bioinformatics-training/2019_March_UCSF_mRNAseq_Workshop/master/scripts/star_stats.sh bash star_stats.sh two. Transfer summary_alignments.txt to your calculator using scp or winSCP, or copy/paste from cat [sometimes doesn't work],
In a new beat session on your laptop. NOT logged into polliwog.
mkdir ~/rnaseq_workshop cd ~/rnaseq_workshop scp msettles@tadpole.genomecenter.ucdavis.edu:/share/workshop/msettles/rnaseq_example/summary_alignments.txt . Its ok of the mkdir control fails ("File exists") because nosotros aleady created the directory before.
Open up in excel (or excel similar application), and lets review.
The table that this script creates ("alignment_stats.txt") can exist pulled to your laptop via 'scp', or WinSCP, etc., and imported into a spreadsheet. Are all samples behaving similarly? Discuss …
Scripts
slurm script for indexing the genome
star_index.slurm
beat script for indexing the genome
star_index.sh
slurm script for mapping using slurm task array and star
star.slurm
beat out script for mapping using bash loop and star.
star.sh
vanquish script to produce summary mapping table
star_stats.sh
welliverhenew1967.blogspot.com
Source: https://ucdavis-bioinformatics-training.github.io/2019_March_UCSF_mRNAseq_Workshop/data_reduction/alignment.html
0 Response to "Igv Number of Reads Aligned to Location"
Enregistrer un commentaire